.. raw:: html
The ``showyourwork.yml`` file
=============================
This is the configuration file for ``showyourwork``, allowing you to
customize several aspects of the workflow. Below is a list of all
available options.
``arxiv_tarball_exclude``
^^^^^^^^^^^^^^^^^^^^^^^^^
**Type:** ``list``
**Description:** List of of files/paths to exclude from the arXiv tarball.
By default, ``showyourwork`` will never include ``python`` scripts, ``matplotlibrc``
config files, ``python`` and ``showyourwork`` temporaries, or ``.gitignore``
files in the tarball. It will also automatically exclude datasets that are
uploaded to/downloaded from Zenodo. This option is useful if there are other
files in your repository -- such as static datasets or other kinds of scripts --
that don't need to be included in the tarball. Note that glob syntax is allowed,
and all paths should be relative to the root of your repository.
**Required:** no
**Default:** ``[]``
**Example:**
.. code-block:: yaml
arxiv_tarball_exclude:
- src/data/dataset.dat
- src/ms.bib
- src/**/*.sh
``CI``
^^^^^^
**Type:** ``bool``
**Description:** Flag indicating whether or not this is a GitHub Actions continuous
integration (CI) build. This is set automatically, but can be overridden here
for debugging purposes.
**Required:** no
**Default:** (inferred automatically)
**Example:**
.. code-block:: yaml
CI: false
``dependencies``
^^^^^^^^^^^^^^^^
**Type:** ``list``
**Description:** List of dependencies for each script. Each entry should be
the path to a script (either a figure script or the TeX manuscript itself)
relative to the repository root. Following each entry, provide a list of
all files on which the script depends. These dependencies may either be
static (such as helper scripts) or programmatically generated (such as
datsets downloaded from Zenodo). In the latter case, instructions on how
to generate them must be provided elsewhere (either via the ``zenodo`` key
below or via a custom ``rule`` in the ``Snakefile``). In both cases, changes
to the dependency will result in a re-run of the section of the workflow that
executes the script.
**Required:** no
**Default:** ``[]``
**Example:**
Tell ``showyourwork`` that the figure script ``my_figure.py`` depends on
a helper script called ``helper_script.py``:
.. code-block:: yaml
dependencies:
src/figures/my_figure.py:
- src/figures/utils/helper_script.py
See :ref:`custom_script_deps`. You can also
specify a dependency on a programmatically-generated file:
.. code-block:: yaml
dependencies:
src/figures/fibonacci.py:
- src/data/fibonacci.dat
See :ref:`custom_dataset_deps`. Finally,
dependencies of the manuscript file are also allowed:
.. code-block:: yaml
dependencies:
src/ms.tex:
- src/answer.tex
See :ref:`custom_ms_deps`.
``download_only``
^^^^^^^^^^^^^^^^^
**Type:** ``bool``
**Description:** If set to ``true``, will never attempt to generate figure
dependencies if they are hosted on Zenodo (instead, showyourwork downloads them).
This behavior is similar to setting ``CI`` to ``true`` and is especially
useful for third-party users who have cloned the repository and don't want
to re-run expensive simulation steps, or don't have the authorization to
upload files to the Zenodo deposit.
**Required:** no
**Default:** ``false``
**Example:**
.. code-block:: yaml
download_only: true
``figexts``
^^^^^^^^^^^
**Type:** ``list``
**Description:** List of recognized figure extensions.
**Required:** no
**Default:** ``[pdf, png, eps, jpg, jpeg, gif, svg, tiff]``
**Example:**
.. code-block:: yaml
figexts:
- pdf
- png
``ms``
^^^^^^
**Type:** ``str``
**Description:** Path to the main TeX manuscript. Change this if you'd prefer to
name your manuscript something other than ``src/ms.tex``. Note that you should still
keep it in the ``src/`` directory. Note also that the compiled PDF will still be named
``ms.pdf`` regardless of this setting.
**Required:** no
**Default:** ``src/ms.tex``
**Example:**
.. code-block:: yaml
ms: src/article.tex
See :ref:`custom_ms_name`.
.. _config_scripts:
``scripts``
^^^^^^^^^^^
**Type:** ``list``
**Description:** List of script extensions and instructions on how to execute
them to generate output. By default, ``showyourwork`` expects output files
(e.g., figures or datasets) to
be generated by executing the corresponding scripts with ``python``. You can add custom
rules here to produce output from scripts with other extensions, or change
the behavior for executing ``python`` scripts (such as adding command line
options, for instance). Each entry under ``scripts`` should be a file extension,
and under each one should be a string specifying how to generate the output file
from the input script. The following placeholders are recognized by ``showyourwork``
and expand as follows at runtime:
- ``{script}``: The full path to the input script.
- ``{script.path}``: The full path to the directory containing the input script.
- ``{script.name}``: The name of the input script (without the path).
- ``{output}``: The full path to the output file.
- ``{output.path}``: The full path to the directory containing the output file.
- ``{output.name}``: The name of the output file (without the path).
**Required:** no
**Default:** The default behavior for ``python`` scripts corresponds to the
following specification in the ``yaml`` file:
.. code-block:: yaml
scripts:
py:
cd {script.path} && python {script.name}
That is, ``python`` is used to execute all scripts that end in ``.py``.
.. important::
By default, ``showyourwork`` always does a ``cd`` into the directory
containing the script and executes it from within that directory; therefore,
any relative paths within ``python`` scripts will be relative to the directory
containing the script.
**Example:**
We can tell ``showyourwork`` how to generate figures from Graphviz ``.gv``
files as follows:
.. code-block:: yaml
scripts:
gv:
dot -Tpdf {script} > {output}
or, to run it from the directory containing the script (as discussed above),
.. code-block:: yaml
scripts:
gv:
cd {script.path} && dot -Tpdf {script.name} > {output}
See :ref:`custom_non_python`.
``tectonic_latest``
^^^^^^^^^^^^^^^^^^^
**Type:** ``bool``
**Description:** Use the latest version of ``tectonic`` (built from source) instead
of the most recent stable version? You shouldn't normally have to edit this entry.
**Required:** no
**Default:** ``false``
**Example:**
.. code-block:: yaml
tectonic_latest: true
``tectonic_os``
^^^^^^^^^^^^^^^
**Type:** ``str``
**Description:** Operating system used for choosing which ``tectonic``
binary to install (only if ``tectonic_latest`` is ``true``).
This is usually determined automatically, but can be
overridden. Options are ``x86_64-unknown-linux-gnu``, ``x86_64-apple-darwin``,
or ``x86_64-pc-windows-msvc``.
**Required:** no
**Default:** (inferred automatically)
**Example:**
.. code-block:: yaml
tectonic_os: x86_64-apple-darwin
``verbose``
^^^^^^^^^^^
**Type:** ``bool``
**Description:** Enable verbose output? Useful for debugging runs.
**Required:** no
**Default:** ``false``
**Example:**
.. code-block:: yaml
verbose: true
.. _zenodo_key:
``zenodo``
^^^^^^^^^^
**Type:** ``list``
**Description:** A list of datasets to be download from and/or uploaded to
Zenodo. Each entry should be the path to a dataset, followed by keys
specifying information about the Zenodo deposit. These keys depend on the use
case. If the deposit already exists (i.e., it was uploaded manually), then
users should only specify the deposit *version* :ref:`id `.
If the deposit does not exist, and users would like ``showyourwork`` to upload
it/download it from Zenodo, they should specify the deposit *concept*
:ref:`id ` instead (see :ref:`id ` below for
more details).
Additionally, users should specify the following keys
(most of which are optional): :ref:`script `,
:ref:`title `,
:ref:`description `,
and :ref:`creators `.
Finally, if the deposit is a tarball consisting of many datasets, users should
also specify the tarball :ref:`contents `.
In both cases (manually uploaded and ``showyourwork``-managed datasets),
a :ref:`token_name ` key is also accepted.
.. note::
For ``showyourwork``-managed datasets, the ``script`` that generates the
dataset will be executed when running the workflow locally (but only if there
are changes to the dataset's dependencies).
When running on GitHub Actions, on the other hand, the script will **never** be
executed; instead, ``showyourwork`` will always download the dataset from
Zenodo. The idea here is to prevent the workflow from executing expensive
operations on the cloud. In order for this to work, however, a deposit must
exist, so you must run your workflow at least once locally before pushing
the changes to GitHub.
**Required:** no
**Default:** ``[]``
**Example:**
See :ref:`custom_dataset_deps`,
:ref:`custom_simulation_deps`,
:ref:`custom_tarballs`,
and
:ref:`custom_tarballs_advanced`.
.. _zenodo.dataset.contents:
``zenodo..contents``
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
**Type:** ``list``
**Description:** If ```` is a ``.tar.gz`` file, users should provide
a list of the contents of the tarball.
If this is a static tarball that was manually uploaded to Zenodo
(i.e., the provided ``id`` is a version id), this should be a list of full
paths to the files to be created when the tarball is extracted.
See below for details.
If, on the other hand, this tarball is managed by
``showyourwork`` (i.e., the provided ``id`` is a concept id), this should be
a list of the full paths of all the files to include in the tarball.
These should be located in the ``src/data`` folder (or nested within it).
Note that instructions for generating these individual files
should be provided separately, either via the :ref:`script `
key or via a custom ``rule`` in the ``Snakefile``.
For static tarballs, users need to be careful when providing file paths.
``showyourwork`` will extract the tarball from the top-level directory of your
repository and attempt to generate all of the files listed in ``contents``,
either by respecting the file path within the tarball or by treating it as a
path relative to the ``src/data`` directory.
For example, consider the Zenodo-hosted file ``results.tar.gz``, whose contents
are
.. code-block::
src/data/results.tar.gz
├── src/data/results_00.dat
└── src/data/results_01.dat
We can specify the following settings for it in ``showyourwork.yml``:
.. code-block:: yaml
zenodo:
- src/data/results.tar.gz:
contents:
- src/data/results_00.dat
- src/data/results_01.dat
which will unpack the files ``results_00.dat`` and ``results_01.dat`` into the
``src/data`` folder. In this case, the source and destination paths are the same
(i.e., the path inside the tarball is the path we extract the files to). But
things will also work if we have a tarball with purely relative paths:
.. code-block::
src/data/other_results.tar.gz
├── other_results_00.dat
└── other_results_01.dat
and we specify the following in ``showyourwork.yml``:
.. code-block:: yaml
zenodo:
- src/data/other_results.tar.gz:
contents:
- src/data/other_results_00.dat
- src/data/other_results_01.dat
In this case, the source and destination paths are different, but ``showyourwork``
knows how to handle it.
Note that if you have files within nested folders inside your tarball, things
should still work as long as you extract them into the ``src/data`` directory.
Note that there is no need to specify the nested directories in ``contents``:
just the full path to the files; intermediate directories will be created as needed.
**Required:** yes, but only if ```` is a ``.tar.gz`` tarball.
**Default:**
**Example:**
See :ref:`custom_tarballs`.
.. _zenodo.dataset.creators:
``zenodo..creators``
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
**Type:** ``list``
**Description:** A list of creators to be listed on the Zenodo record and associated
with the record DOI.
**Required:** no
**Default:** The GitHub username of the current user
**Example:**
See :ref:`custom_simulation_deps`.
.. _zenodo.dataset.description:
``zenodo..description``
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
**Type:** ``str``
**Description:** A detailed description of the file, how it was generated, and
how it should be used, to be displayed on the Zenodo record page.
**Required:** no
**Default:** ``"File uploaded from "``
**Example:**
See :ref:`custom_simulation_deps`.
.. _zenodo.dataset.id:
``zenodo..id``
^^^^^^^^^^^^^^^^^^^^^^^
**Type:** ``int``
**Description:** A Zenodo ``id`` for a given deposit is the last part of its DOI. For example,
a deposit with DOI ``10.5281/zenodo.5749987`` has ``id`` equal to ``5749987``.
This is also the last part of the url for the corresponding record
(``_). Importantly, Zenodo makes a distinction
between *version* DOIs and *concept* DOIs. Version DOIs are static, and tied
to a specific version of a deposit (the way you'd expect a DOI to behave).
This is the type of ``id`` you should provide if you manually uploaded a dataset
to Zenodo and only ever want ``showyourwork`` to download it.
Concept DOIs, on the other hand, point to *all* versions of a given record,
and always resolve to the *latest* version. If you want ``showyourwork``
to manage the dataset for you by generating it, uploading it, and downloading
it, this is the kind of ``id`` you should provide.
Check out the sidebar on the
`web page for the deposit in the example above `_:
.. raw:: html
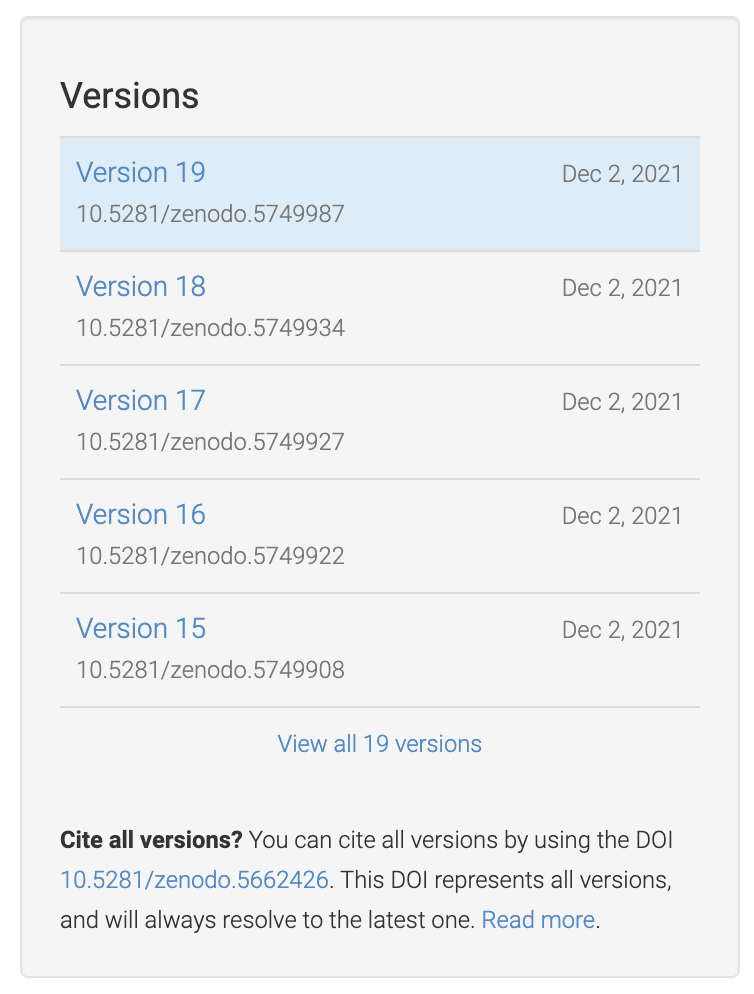
You can see that the ``id`` ``5749987`` corresponds to a specific version (``19``)
of the deposit, while the ``id`` ``5662426`` corresponds to *all* versions of
the deposit (it's listed under "Cite all versions?").
The former is a "version" id, while the latter is a "concept" id.
You can read more about that in the `Zenodo docs `_.
.. note::
If you're just getting started and want a concept ``id`` for a fresh draft
of a new Zenodo deposit, run
.. code-block:: bash
make reserve
.. raw:: html
from the top level of your repo. This will pre-reserve a concept ``id`` for
you (assuming you're properly authenticated) and print it to the terminal.
**Required:** yes
**Default:**
**Example:**
The following snippet
.. code-block:: yaml
zenodo:
- src/data/results.tar.gz:
id: 5749987
tells ``showyourwork`` to download the file ``results.tar.gz`` from
the static Zenodo deposit at ``_
(version 19 of the deposit, as mentioned above). This file must already
exist, and ``showyourwork`` won't ever attempt to re-generate it or
re-upload it to Zenodo because it recognizes ``5749987`` as a *version* id.
.. raw:: html
Alternatively, we could specify the following:
.. code-block:: yaml
zenodo:
- src/data/results.tar.gz:
id: 5662426
script: src/analysis/generate_results.py
In this case, the ``id`` is a *concept* id, corresponding to all versions of
the deposit, and ``showyourwork`` will take over management of the deposit.
Note that we also provided a ``script`` instructing ``showyourwork`` how to
generate new versions of the deposit. Whenever ``generate_results.py`` or
any of its dependencies are modified, ``showyourwork`` will re-generate
``results.tar.gz`` **and re-upload it to Zenodo under the same concept id**
when running the workflow locally.
This will create a new version DOI under the same concept DOI.
Note that in order for this to work, you must be properly authenticated;
see :ref:`token_name ` below.
For a more detailed example, see :ref:`custom_dataset_deps`.
.. _zenodo.dataset.script:
``zenodo..script``
^^^^^^^^^^^^^^^^^^^^^^^^^^^
**Type:** ``str``
**Description:** The path to the ``python`` script that generates the ````
(or, if ```` is a tarball, the script that generates its contents).
Note that this *must* be a ``python`` script, even if custom script instructions
are provided via the :ref:`scripts ` key. To define custom rules for
generating the dataset, see the
:ref:`custom_tarballs_advanced` example.
**Required:** yes, unless a custom ``rule`` is provided in the ``Snakefile``
**Default:**
**Example:**
See :ref:`custom_simulation_deps`.
.. _zenodo.dataset.title:
``zenodo..title``
^^^^^^^^^^^^^^^^^^^^^^^^^^
**Type:** ``str``
**Description:** The title of the Zenodo deposit.
**Required:** no
**Default:** ``":"``
**Example:**
See :ref:`custom_simulation_deps`.
.. _zenodo.dataset.token_name:
``zenodo..token_name``
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
**Type:** ``str``
**Description:** The name of the environment variable containing the
Zenodo access token.
To obtain this token, create a `Zenodo account `_
(if you don't have one already) and
generate a `personal access token `_.
Make sure to give it at least ``deposit:actions`` and ``deposit:write`` scopes, and store it somewhere
safe. Then, assign your token to an environment variable called ``ZENODO_TOKEN`` (or whatever
you set ``token_name`` to). I export mine from within my ``.zshrc`` or ``.bashrc`` config file so that
it's always available in all terminals.
.. warning::
Never include your personal access tokens in any files committed to GitHub!
**Required:** no
**Default:** ``ZENODO_TOKEN``
**Example:**
See :ref:`custom_simulation_deps`.
``zenodo_sandbox``
^^^^^^^^^^^^^^^^^^
**Type:** ``list``
**Description:** A list of datasets to be download from and/or uploaded to
Zenodo Sandbox. This key behaves in the same way and accepts all the same
arguments as the :ref:`zenodo ` key above, but it interfaces with
``_ (instead of ``_). Zenodo Sandbox works in
the same way as Zenodo, but is meant for testing purposes only: deposits hosted
in the Sandbox may be deleted at any time. Hosting datasets here is useful
during development of your project; just make sure to switch over to
``zenodo`` when you're ready to publish your paper!
**Required:** no
**Default:** ``[]``
**Example:**
See :ref:`custom_dataset_deps`,
:ref:`custom_simulation_deps`,
:ref:`custom_tarballs`,
and
:ref:`custom_tarballs_advanced`.